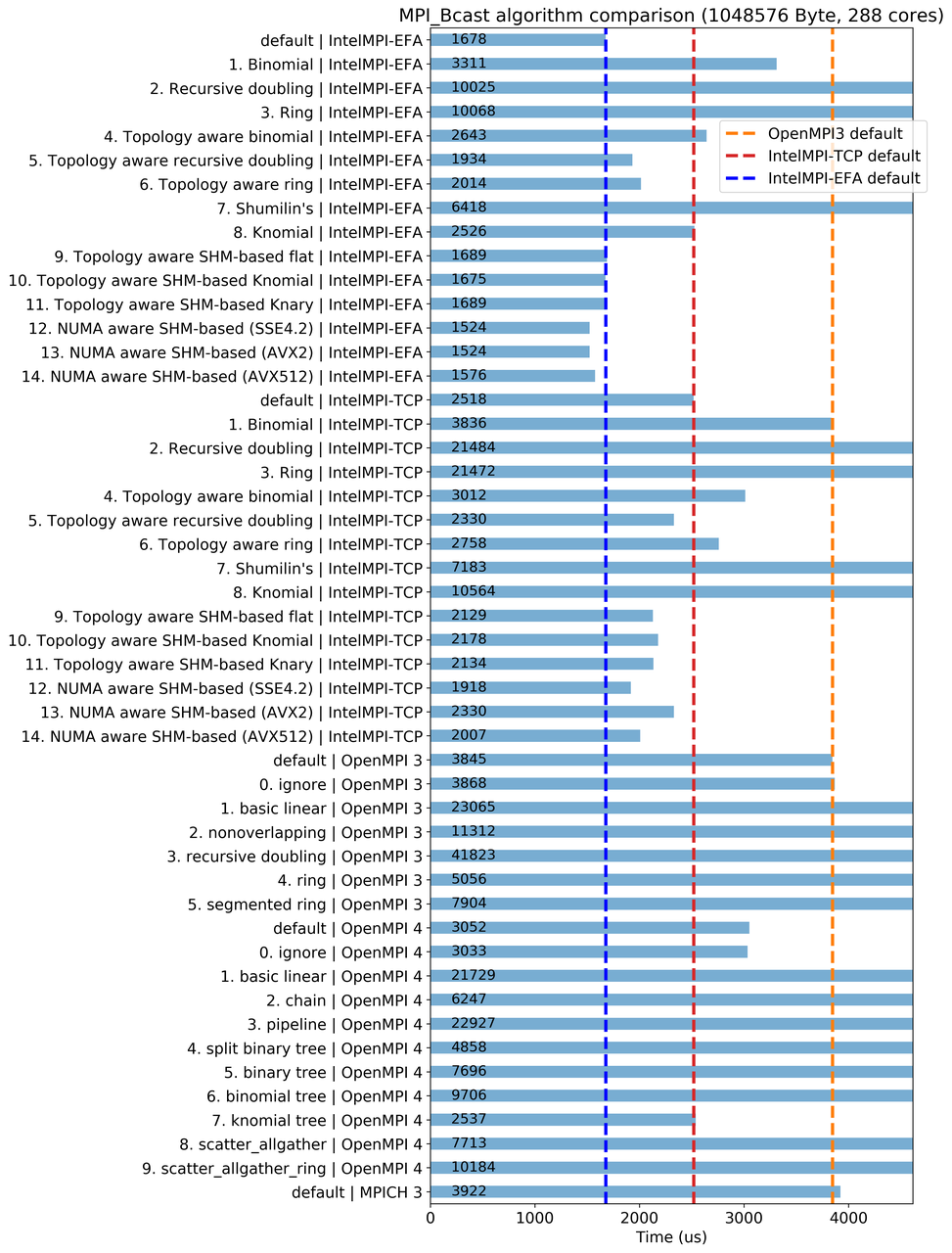
I decide to build my new website and occasionally post blogs on research, software, and computing.
For the blogging tool, basic requirements are:
It must be a static site that can be easily version-controlled on GitHub. So, not WordPress.
I'd like the framework to be written in Python. So, not Jekyll.
It should support Jupyter notebook format, so I can easily post computing & data analysis results.
The options come down to Pelican (the most popular one), Tinkerer (Sphinx-extended-for-blog), and Nikola. I finally settle down on Nikola because it is super intuitive, has a clear and user-friendly documentation, and supports Jupyter natively. It takes me like 10 minutes to set up the desired website layout. Other tools are also very powerful but it takes me longer to get the configuration right.
This post records my setup steps, to help others and the forgetful future me. Building such a website only requires basic knowledge in:
I assume no knowledge in Web front-end stuff (HTML/CSS/JS). This should be typical for many computational & data science students.
Just by Googling you can find tons of tutorials on this. Particularly good ones are:
However this post is still useful because I:
use nikola v8.0.1 as of Feb 2019. Many early tweaks are not necessary now.
focus on personal website (with additional pages like Bio), not only a blog.
add additional notes on GitHub deployment and version control.
The official getting-started guide explains this well. I just summarize the steps here.
I come from the scientific side of Python, so use Conda to create a new environment:
conda create -n blog python=3.6
source activate blog
pip install Nikola[extras]
Making the first demo site is as simple as
Run nikola init in a new folder, with default settings.
Then run nikola auto to build the page and start a local server for testing.
Visit localhost:8000 in the web browser.
Adding a new blog post is as simple as:
Run nikola new_post (defaults to reStructuredText format, add -f markdown for *.md format)
Write blog contents in the newly generated file (*.rst or *.md) in the posts/ directory.
Rerun nikola auto. You can also keep this command running so new contents will be automatically updated.
All configurations are managed by a single conf.py file, which is extremely neat.
Before adding any real contents, you should first configure GitHub deployment, layout, themes, etc.
There is an official deployment guide but I feel that more explanation is necessary.
Most posts mention deployment at the final step, but I think it is good to do so at the very beginning.
We will use GitHub pages to host the website freely. Any GitHub repo can have its own GitHub page, but we will use the special one called user page that only works for a repo named [username].github.io, where [username] is your GitHub account name (mine is jiaweizhuang). The website URL will be https://[username].github.io/.
You will need to:
Create a new GitHub repo named [username].github.io.
Initialize a Git repo (git init) in your Nikola project directory.
Link to your remote GitHub repo (git remote add origin https://github.com/[username]/[username].github.io.git)
So far all standard git practices. The non-standard thing is that you shouldn't manually commit anything to the master branch. The master branch is used for storing HTML files to display on the web. It should be handled automatically by the command nikola github_deploy. To version control your source files (conf.py, *.rst, *.md), you should create a new branch called src:
git checkout -b src
My .gitignore is:
cache
.doit.db*
__pycache__
output
.ipynb_checkpoints
.DS_Store
You can manually commit to this src branch and push to GitHub.
In conf.py, double-check that branch names are correct:
GITHUB_SOURCE_BRANCH = 'src'
GITHUB_DEPLOY_BRANCH = 'master'
GITHUB_REMOTE_NAME = 'origin'
I also recommend setting:
GITHUB_COMMIT_SOURCE = False
So that the nikola github_deploy command below won't touch your src branch.
To deploy the content on master, run:
nikola github_deploy
This builds the HTML files, commits to the master branch, and pushes to GitHub. The actual website https://[username].github.io/ will be updated in a few seconds.
You end up having:
A well version-controlled src branch, with only source files. You can add meaningful commit messages like for other code projects.
An automatically generated master branch, with messy html files which you never need to directly look at. It doesn't have meaningful commit messages, and the commit history is kind of a mess (diff between HTML files).
Note
Remember that nikola github_deploy will use all the files in the current directory, not the most recent commit in the src branch! master and the src are not necessarily synchronized if you set GITHUB_COMMIT_SOURCE = False.
For all the tweaks later, you can incrementally update the GitHub repo and the website, by manually pushing to src and using nikola github_deploy to push to master.
The default theme looks more like a blog than a personal website. Twitter's Bootstrap is an excellent theme and is built into Nikola. In conf.py, set:
THEME = "bootstrap4"
The theme can be further tweaked by Bootswatch but I find the default theme perfect for me :)
Official non-blog guide explains this well. I just summarize the steps here.
Nikola defines two types of contents:
"Posts" generated by nikola new_post. It is just the blog post and will be automatically added to the main web page whenever a new post is created.
"Pages" generated by nikola new_page. It is a standalone page that will not be automatically added to the main site. This is the building block for a non-blog site.
In conf.py, bring Pages to root level:
POSTS = (
("posts/*.rst", "blog", "post.tmpl"),
("posts/*.md", "blog", "post.tmpl"),
("posts/*.txt", "blog", "post.tmpl"),
("posts/*.html", "blog", "post.tmpl"),
)
PAGES = (
("pages/*.rst", "", "page.tmpl"), # notice the second argument
("pages/*.md", "", "page.tmpl"),
("pages/*.txt", "", "page.tmpl"),
("pages/*.html", "", "page.tmpl"),
)
INDEX_PATH = "blog"
Generate your new index page (the entry of you website):
$ nikola new_page
Creating New Page
-----------------
Title: index
To add more pages to the top navigation bar:
$ nikola new_page
Creating New Page
-----------------
Title: Bio
And then add it to conf.py:
NAVIGATION_LINKS = {
DEFAULT_LANG: (
("/index.html", "Home"),
("/bio/index.html", "Bio"),
...
),
For *.rst posts, simple add:
.. contents::
Or optionally with numbering:
.. contents::
.. section-numbering::